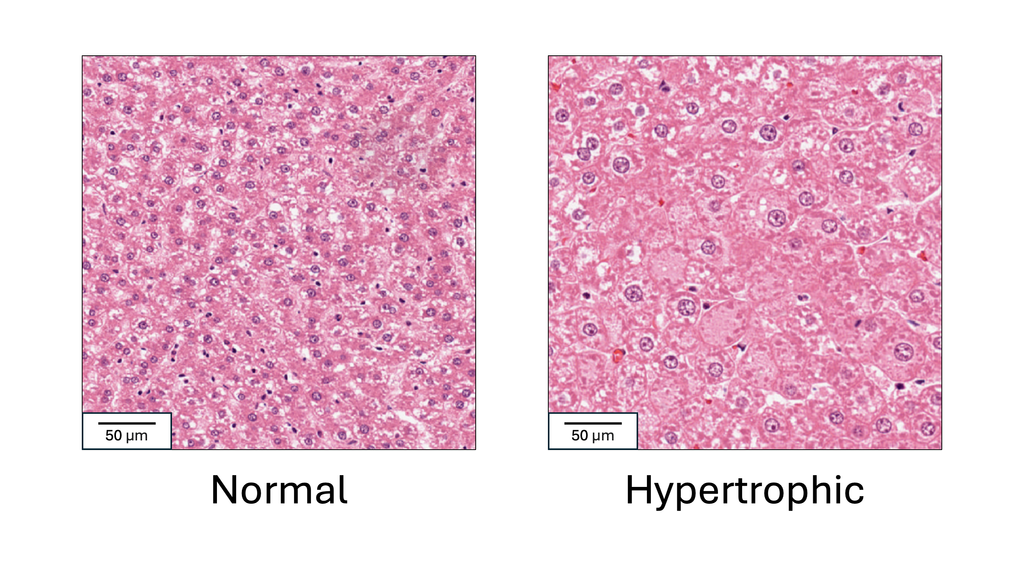
Problem
Drug safety studies are a key step in drug development and approval. In these studies, a group of experimental animals is given the target drug, and a control group is given a placebo or vehicle substance. Tissue samples from both groups are then microscopically analyzed by a pathologist to determine if any abnormalities are present in the target group. Hypertrophy, or cell enlargement, is a critical abnormality to detect in these studies, as it can indicate organ dysfunction, hormonal disruption or early carcinogenic effects. It often occurs in the liver, where normal cell size varies with age, strain, and diet, making it difficult to establish a universal threshold for hypertrophy. Therefore, to determine its presence, the pathologist compares tissues from the treated animals to those of the control animals. This is a challenging and highly time-consuming task, and the need for comparison can lead to missing hypertrophy when it is subtle. In this project, we aim to automatically detect liver hypertrophy on digital histology slides to reduce the time and workload associated with drug development, which could allow drugs to reach those who need them sooner. We also aim to reduce the rate of missed hypertrophy, helping to prevent toxic drugs from advancing in the development pipeline.
Approach
Several studies have demonstrated that AI models can successfully detect liver hypertrophy in drug safety studies [1-5]. The most accurate study (AUC > 0.95) was also the only one to use a state-of-the-art foundation model [2]. However, this study was based on a relatively small dataset (n=29 slides with hypertrophy), making it difficult to determine whether the foundation model approach is truly optimal for this problem. The first goal of this project is therefore to evaluate whether existing foundation models can achieve comparable performance on a larger and more diverse dataset.
Additionally, all prior approaches have analyzed individual slides in isolation, whereas pathologists rely on comparative analysis across multiple slides to detect hypertrophy. This single-slide approach may be prone to errors due to background variability. The second goal of this project is to enhance existing models —or develop a novel one— to incorporate control group data, thereby aligning more closely with human pathology workflows and potentially improving detection accuracy.
Data
The data for this project come from TG-GATES, a publicly available repository of rat drug studies [6]. TG-GATES contains over 23,000 liver digital pathology slides, organized by drug study, with labels indicating whether each slide belongs to the control or treatment group and whether the slide has hypertrophy. Among these, more than 1,000 slides exhibit hypertrophy. A local copy of this repository is available.
References
[1] Bertani V, Blanck O, Guignard D, Schorsch F, Pischon H (2021). Artificial Intelligence in Toxicological Pathology: Quantitative evaluation of Compound-Induced follicular cell hypertrophy in rat thyroid gland using deep learning models. Toxicologic Pathology. 50: 23–34.
[2] Cheng Z, Dai W, Sun J (2024). Visual Language model for Preclinical Toxicologic liver histopathology assessment. VLM4Bio’24. 41–48. - doi: 10.1145/3689096.3689463.
[3] Kuklyte J, Fitzgerald J, Nelissen S, Wei H, Whelan A, Power A, Ahmad A, Miarka M, Gregson M, Maxwell M, Raji R, Lenihan J, Finn-Moloney E, Rafferty M, Cary M, Barale-Thomas E, O’Shea D (2021). Evaluation of the use of Single- and Multi-Magnification convolutional neural networks for the determination and quantitation of lesions in nonclinical pathology studies. Toxicologic Pathology. 49: 815–842.
[4] Pischon H, Mason D, Lawrenz B, Blanck O, Frisk A-L, Schorsch F, Bertani V (2021). Artificial intelligence in Toxicologic Pathology: Quantitative evaluation of Compound-Induced hepatocellular hypertrophy in rats. Toxicologic Pathology. 49: 928–937.
[5] Shimazaki T, Deshpande A, Hajra A, Thomas T, Muta K, Yamada N, Yasui Y, Shoda T (2021). Deep learning-based image-analysis algorithm for classification and quantification of multiple histopathological lesions in rat liver. Journal of Toxicologic Pathology. 35: 135–147.
[6] Igarashi Y, Nakatsu N, Yamashita T, Ono A, Ohno Y, Urushidani T, Yamada H (2015). Open TG-GATEs: a large-scale toxicogenomics database. Nucleic Acids Research. 43: D921–D927.
Requirements
- Students with a major in computer science, biomedical engineering, artificial intelligence, physics, or a related area in the final stage of master level studies are invited to apply.
- Affinity with programming in Python, specifically in the PyTorch framework.
- Interest in deep learning and medical image analysis.
Information
- Project duration: 6-9 months
- Location: Radboud University Medical Center
- You will be part of the Diagnostic Image Analysis Group (DIAG) and Computational Pathology Group, whose research focus is the analysis of histopathological slides with deep learning techniques.
- You will have access to and work with a large GPU cluster.
- For more information, please contact Salma Dammak.

