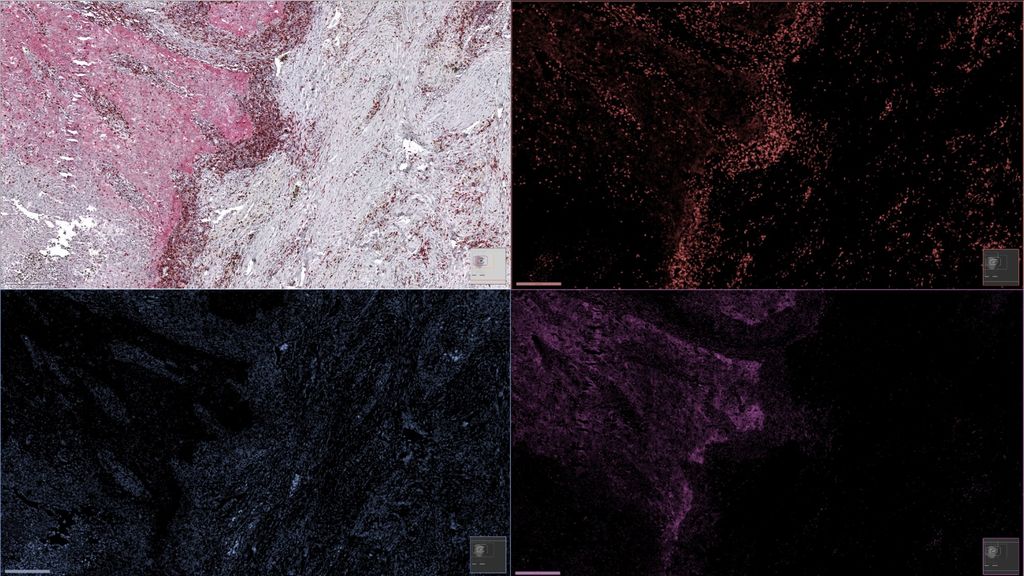
Background
Immunohistochemical (IHC) staining is a commonly used technique in pathology to target specific types of cells with specific chromogens, to help pathologists recognize different types of cells in tissue samples of breast cancer, for example. When pathologists are interested in multiple types of cells, multiple IHC markers are needed, which are usually applied to different (consecutive) tissue sections. Multiplex immunohistochemistry (mIHC) is a technique that allows staining a single slide with multiple IHC markers, using multiple colors. In this way, the spatial tissue architecture, the interaction of different types of cells present on the same slide, and the positivity of some cells to multiple markers at the same time, can be analyzed. But in order to analyze each marker independently, for example using computer techniques of advanced image analysis after slide digitization, we need to unmix the multiple colors, which is a challenging problem, especially when the number of colors is fairly large.
Project goal
Develop deep learning models to automatically unmix digital pathology slides stained with mIHC. Possible techniques are (not limited to) unsupervised models such as convolutional autoencoders. Optionally, develop a simple (Python-based) user interface to calibrate the software by, for example, manually selecting different image color regions.
Requirements
- Students with a major in computer science, biomedical engineering, artificial intelligence, physics, or a related area in the final stage of master level studies are invited to apply.
- Affinity with programming in Python
- Interest in deep learning and medical image analysis
Information
- Project duration: 6 months
- Location: Radboud University Nijmegen Medical Center
- For more information please contact Witali Aswolinskiy or Francesco Ciompi

