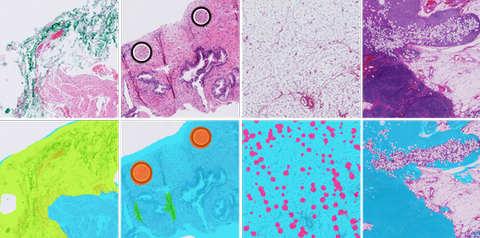
Clinical Problem
In an AI-powered digital pathology workflow, the process of going from glass slide to digitization and analysis with artificial intelligence suffers from two main problems.
First, the quality of the slide needs to be assessed right after, or even during, scanning, to guide decisions such as rescanning the slide (e.g., if partly out of focus), further cleaning the glass, or producing a new tissue slide.
Second, after scanning, to identify areas to be optionally flagged as non-relevant (e.g., adipose tissue) or that can substantially affect model performance (e.g., folded tissue, ink, crushed artifacts), which can be weighed during the analysis done by a downstream AI algorithm.
When analyzing whole slide images, identifying tissue areas and removing the background is a relevant step to prevent unnecessary processing of large parts of the WSI[1]. Moreover, the digitization process can lead to image artifacts, such as out-of-focus, tissue folds, ink, dust, pen marks and air bubbles.
Artifacts can obstruct tissue parts and significantly affect model performance, which means that a quality control mechanism is required to rule out poor quality slides or defective tissue regions.
Preliminary work in our group has resulted in promising results, but model robustness across different stainings, especially immunohistochemical markers, needs to be improved[2]. Depending on the specific task, we may also wish to detect and exclude other regions of whole-slide images such as adipose tissue, small cores or control samples in IHC. One of the biggest challenges in digital pathology comes from having different stains, tissue thicknesses and scanners and the purpose of AI algorithms is to be robust enough to perform well with vastly different images.
[1] Bándi et al, Resolution-agnostic tissue segmentation in whole-slide histopathology images with convolutional neural networks
[2] Smit et al, Quality control of whole-slide images through multi-class semantic segmentation of artifacts
Solution
The goals of this project include the development of multi-task artificial intelligence algorithms to analyze whole-slide images and perform quality control tasks, such as separating tissue from background across a variety of stainings, detecting artifacts, non-clinically relevant regions of the slide such as the ones containing adipose tissue, and control tissue samples in immunohistochemistry, which need to be distinguished from the target tissue in the slide. Given the amount of tasks to perform and the need for fast quality control and pre-processing in the digital pathology workflow, we aim at developing efficient deep learning models able to perform fast inference. The main result of the project will be an artificial intelligence algorithm with source code and documentation; the algorithm will be made publicly available as a Docker container on https://grand-challenge. org. The docker should have different modes for background segmentation only, artifact detection, adipose tissue segmentation, etc. Optionally, developed AI models can be ported to an edge compute device (e.g., the Intel Neural Compute Stick, https://www.intel.com/content/www/us/en/developer/tools/neural-compute-stick/overview.html), to be used as a proof of concept of optimized fast inference approach towards adoption in the clinic, placed as close as possible to the digital pathology scanner, without the need for additional layers of data transfer, such as cloud computing.
Data
In this project we intend to combine external and inhouse dataset of whole-slide images consisting of different tissues and stain types, including manual annotations.
Approach
The student will be supervised by a research member of the Diagnostic Image Analysis Group and Computational Pathology Group whose research is dedicated to analyses of histopathological slides with deep learning techniques. The student will have access to a large GPU cluster.
Requirements
- Students with a major in computer science, biomedical engineering, artificial intelligence, physics, or a related area in the final stage of master level studies are invited to apply;
- affinity with programming in Python;
- interest in deep learning and medical image analysis.
Information
- Project duration: 6 months
- Location: Radboud University Medical Center
- For more information, please contact Marina D’Amato

